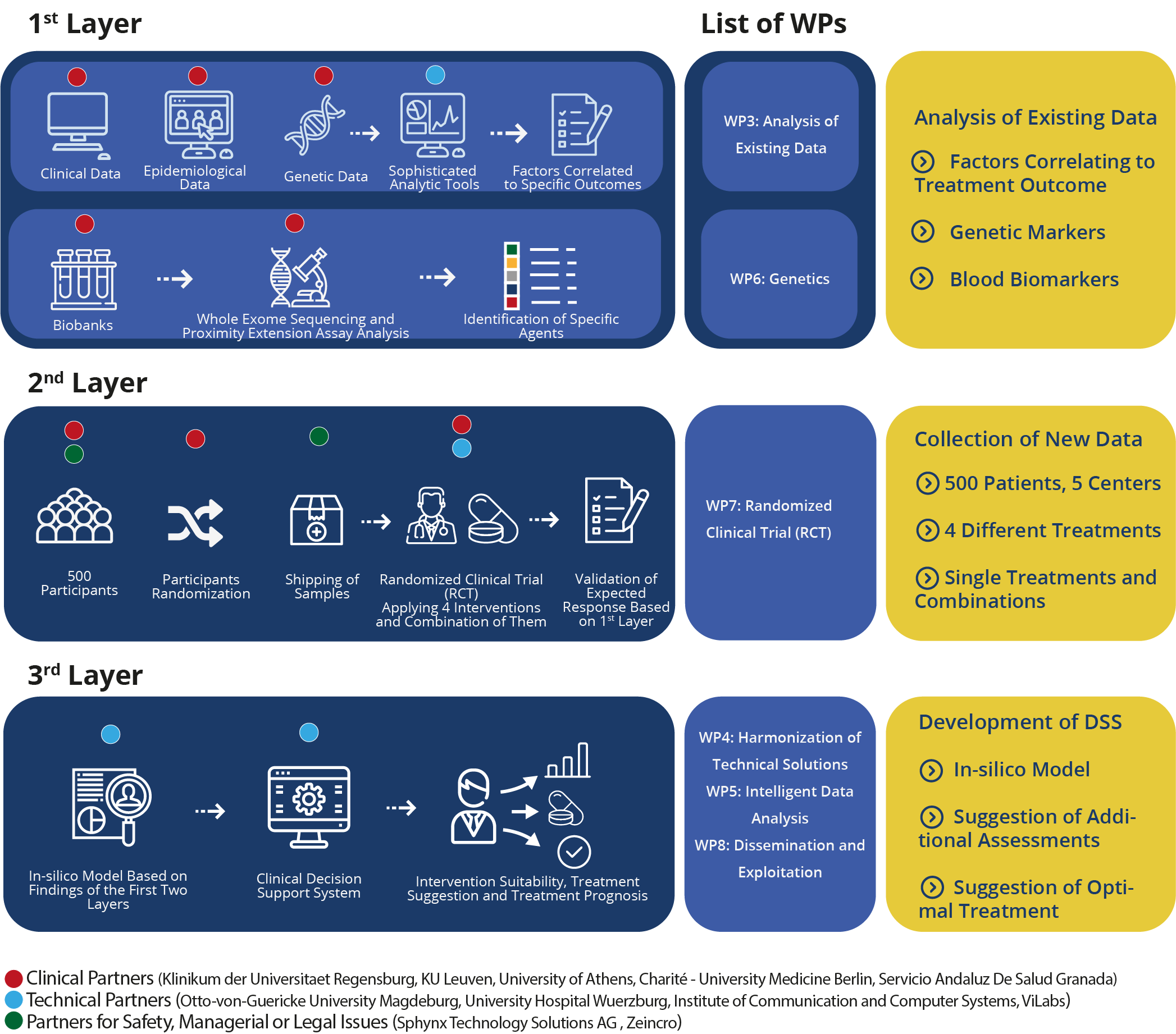
UNITI can be perceived as a three-layered project:
In the first layer, existing clinical, epidemiological and genetic/blood biomarker data will be re-analysed with sophisticated analytic tools in order to identify factors correlated to specific treatment outcomes in the individual patient. Moreover, GRA and KI will perform additional analysis of their biobanks aiming towards the identification of candidate genes (rare and common variants), using Whole Exome Sequencing (WES) techniques. Additionally, an attempt to identify blood biomarkers specific to clinical subtypes, will be performed using a panel of the Proximity Extension Assay (PEA) method.
In the second layer, a large scale multicentre RCT will be conducted, with use of additional dimensions of analysis. Aim of this trial will not be only the comparison of tinnitus interventions’ efficacy but also the validation of the treatment outcome predictions of the DSS.
The third layer will be the construction of an easy to use clinical DSS, which, based on the findings of the first two layers, will be able to identify the suitability of the particular patient for a list of the available interventions. More specifically, it will provide suggestion for specific examinations considered valuable for prediction of optimal treatment or combination of treatments, based on how good patient’s profile fits to the identified patterns. DSS will be a component of the UNITI platform along with the in-silico model (OBJ7) and the mobile application (OBJ3).